R Basics
2025-09-15
Packages
R has a base installation and then tens of thousands of add-on software packages that can be obtained from CRAN
Use
install.packagesto install the dslabs package from CRANTry out the following functions:
sessionInfo,installed.packages
Prebuilt functions
Much of what we do in R uses functions from either base R or installed packages.
Many are included in automatically loaded packages:
stats,graphics,grDevices,utils,datasets,methods.This subset of the R universe is referred to as R base.
Very popular packages not included in R base:
ggplot2,dplyr,tidyr, anddata.table.It is easy to write your own functions and packages
Important
For problem set 2 you can only use R base.
Base R functions
Example functions that we will use today:
ls,rm,library,search,factor,list,exists,str,typeof, andclass.You can see the raw code for a function by typing its name without the parentheses.
- type
lson your console to see an example.
- type
Help system
In R you can use
?orhelpto learn more about functions.You can learn about function using
or
- many packages provide vignettes that are like how-to manuals that can show how the functions in the package are meant to be used to create different analyses
The workspace
- Define a variable.
- Use
lsto see if it’s there. Also take a look at the Environment tab in RStudio.
- Use
rmto remove the variable you defined.
The workspace
each time you start R you will get a new workspace that does not have any variables or libraries loaded
when you quit R you will be asked if you want to save the workspace
- you will probably always be better off saying NO
if you do save a workspace it will be saved as a hidden file (Unix lecture) and whenever R is started in a directory with a saved workspace then that workspace will be re-instantiated and used
- this might be helpful if you are working on a long complex analysis with large data
mostly it is much easier to just use a markdown document to detail your steps and rerun them on a clean workspace
- this can also help find errors in your analysis
The Workspace
- just like Unix R uses a search path to find functions that you can evaluate
Variable name convention
A nice convention to follow is to use meaningful words that describe what is stored, only lower case, and underscores as a substitute for spaces.
R and RStudio both provide autocomplete capabilities so you don’t need to type the whole name
- this makes it easier to use longer, more descriptive variable names
It is highly recommended that you not use the period
.in variable names, there are situations where R treats it differently and those can cause unintended actionsFor more we recommend this guide.
Data types
The main data types in R are:
One dimensional vectors: double, integer, logical, complex, characters.
integer and double are both numeric
Factors
Lists: this includes data frames.
Arrays: Matrices are the most widely used.
Date and time
tibble
S4 objects
Data types
Many errors in R come from confusing data types.
strstands for structure, gives us information about an object.typeofgives you the basic data type of the object. It reveals the lower-level, more fundamental type of an object in R’s memory.classThis function returns the class attribute of an object. The class of an object is essentiallytype_ofat a higher, often user-facing level.
Data types
Let’s see some example:
[1] "list"[1] "data.frame"'data.frame': 51 obs. of 5 variables:
$ state : chr "Alabama" "Alaska" "Arizona" "Arkansas" ...
$ abb : chr "AL" "AK" "AZ" "AR" ...
$ region : Factor w/ 4 levels "Northeast","South",..: 2 4 4 2 4 4 1 2 2 2 ...
$ population: num 4779736 710231 6392017 2915918 37253956 ...
$ total : num 135 19 232 93 1257 ...[1] 51 5Data frames
Date frames are the most common class used in data analysis.
Data frames are like a matrix, but where the columns can have different types.
Usually, rows represents observations and columns variables.
you can index them like you would a matrix,
x[i, j]refers to the element in rowicolumnjYou can see part of the content like this
Data frames
- and you can use
Viewto open a spreadsheet-like interface to see the entire data.frame.
- This is more effective in RStudio because it has an integrated viewer.
Data frames
- A very common operation is adding columns like this:
state abb region population total pop_rank
1 Alabama AL South 4779736 135 29
2 Alaska AK West 710231 19 5
3 Arizona AZ West 6392017 232 36
4 Arkansas AR South 2915918 93 20
5 California CA West 37253956 1257 51
6 Colorado CO West 5029196 65 30Data frames
Note that we used the
$.This is called an
accessorbecause it lets us access columns.
[1] 4779736 710231 6392017 2915918 37253956 5029196 3574097 897934
[9] 601723 19687653 9920000 1360301 1567582 12830632 6483802 3046355
[17] 2853118 4339367 4533372 1328361 5773552 6547629 9883640 5303925
[25] 2967297 5988927 989415 1826341 2700551 1316470 8791894 2059179
[33] 19378102 9535483 672591 11536504 3751351 3831074 12702379 1052567
[41] 4625364 814180 6346105 25145561 2763885 625741 8001024 6724540
[49] 1852994 5686986 563626- More generally:
$can be used to access named components of a list.
Data frames
One way R confuses beginners is by having multiple ways of doing the same thing.
For example you can access the 4th column in the following five different ways:
- In general, we recommend using the name rather than the number as adding or removing columns will change index values, but not names.
with
withlet’s us use the column names as symbols to access the data.This is convenient to avoid typing the data frame name over and over:
with
- Note you can write entire code chunks by enclosing it in curly brackets:
Vectors
The columns of data frames are an example of one dimensional (atomic) vectors.
An atomic vector is a vector where every element must be the same type.
Vectors
Often we have to create vectors.
The concatenate function
cis the most basic way used to create vectors:
- We access the elements using
[]
- NOTE R does not do array bounds checking…it silently pads with missing values
Sequences
- Sequences are a common example of vectors we generate.
- When you want a sequence that increases by 1 you can use the colon
:
Sequences
- A useful function to quickly generate a sequence the same length as a vector
xisseq_along(x)and NOT1:length(x)
- A reason to use this is to loop through entries:
- But if the length of
xis zero, then using1:length(x)does not work for this loop
Vector types and coercion
One dimensional vectors: double, integer, logical, complex, characters and numeric
Each basic type has its own version of NA (a missing value)
testing for types:
is.TYPE,coercing :
as.TYPE, will result in NA if it is not possible
Coercing
When you do something inconsistent with data types, R tries to figure out what you mean and change it accordingly.
We call this coercing.
R does not return an error and in some cases does not return a warning either.
This can cause confusion and unnoticed errors.
So it’s important to understand how and when it happens.
Vector types and coercion
coercion is automatically performed when it is possible
TRUEcoerces to 1 andFALSEto 0but any non-zero integer coerces to TRUE, only 0 coerces to FALSE
as.logicalconverts 0 and only 0 toFALSE, everything else toTRUEthe character string “NA” is not a missing value
Coercing
- Here are some examples:
Coercing
- When R can’t figure out how to coerce, rather an error it returns an NA:
- Note that including
NAs in arithmetical operations usually returns anNA.
Coercing
You can coerce explicitly
Most coercion functions start with
as.Here is an example.
Coercing
- The
readrpackage provides some tools for trying to par
Factors
One key distinction between data types you need to understad is the difference between factors and characters.
The
murderdataset has examples of both.
- Why do you think this is?
Factors
A factor is a good representation for a variable, that has a fixed set of non-numeric values
- ex: Sex has
MaleandFemale
- ex: Sex has
It is usually not a good representation for variables that have lots of levels (like state names in the murders dataset)
Internally a factor is stored as the unique set of labels (called levels) and an integer vector with values in 1 to
length(levels)- if the
ith entry iskthen that corresponds to thekth element of the levels
- if the
In statistics we refer to this as categorical data, where all of the individuals are mapped into a relatively small number of categories.
Usually order does not matter, but if it does you can also have ordered factors.
Setting Levels
you can set up the levels as you would like, when creating a factor
if you do not set them up, then they will be created in lexicographic order (in the locale you are using)
x = sample(c("Male", "Female"), 50, replace =TRUE)
y1 = factor(x, levels=c("Male", "Female"))
y2 = factor(x, levels = c("Female", "Male"))
y1[1:10] [1] Male Male Female Female Female Female Female Male Female Male
Levels: Male Female [1] Male Male Female Female Female Female Female Male Female Male
Levels: Female MaleCategories based on strata
In data analysis we often want to stratify continuous variables into categories.
The function
cuthelps us do this:In this case there may be a reason to think of using ordered factors.
Categories based on strata
- We can assign more meaningful level names:
age <- c(5, 93, 18, 102, 14, 22, 45, 65, 67, 25, 30, 16, 21)
cut(age, c(0, 11, 27, 43, 59, 78, 96, Inf),
labels = c("Alpha", "Zoomer", "Millennial", "X", "Boomer", "Silent", "Greatest")) [1] Alpha Silent Zoomer Greatest Zoomer Zoomer
[7] X Boomer Boomer Zoomer Millennial Zoomer
[13] Zoomer
Levels: Alpha Zoomer Millennial X Boomer Silent GreatestChanging levels
This is often needed for ordinal data because R defaults to alphabetical order
Or as noted you may want to make use of
orderedfactors
- You can change this with the
levelsargument:
Changing levels
A common reason we want to change levels is to assure R is aware which is the reference strata.
This is important for linear models because the first level is assumed to be the reference.
Changing levels
We often want to order strata based on a summary statistic.
This is common in data visualization.
We can use
reorderfor this:
Factors
- Another reason we used factors is because they are stored more efficiently:
80000232 bytes40000648 bytes- An integer uses less memory than a character string (but it is a bit more complicated)
Factors can be confusing
- Try to make sense of this:
Factors can be confusing
- Avoid keeping extra levels with
droplevels:
- But note what happens if we change to another level:
NAs
NA stands for not available and represents data that are missing.
Data analysts have to deal with NAs often.
In R there is a different kind of NA for each of the basic vector data types.
There is also the concept of NULL, which represents a zero length list and is often returned by functions or expressions that do not have a specified return value
NAs
- dslabs includes an example dataset with NAs
- The
is.nafunction is key for dealing with NAs
NAs
- Caution logical operators like and (
&) and or (|) coerce their arguments when needed and possible - the logical operators evaluate arguments in a “lazy” fashion, and left to right
NaNs and Inf
A related constant is
NaNwhich stands for Not a NumberNaNis a double, coercing it to integer yields an NAInf and -Inf represent values of infinity and minus infinity (RStudio makes using these really annoying)
Lists
Data frames are a type of list.
Lists permit components of different types and, unlike data frames, different lengths:
- The JSON format is best represented as list in R.
Lists
- You can access components in different ways:
Matrices
Matrices are another widely used data type.
They are similar to data frames except all entries need to be of the same type.
We will learn more about matrices in the High Dimensional Data Analysis part of the class.
Functions
- You can define your own function. The form is like this:
the values you pass to the function are called the arguments and they can have default values (e.g above z is 0 unless provided)
arguments are matched by either name (which takes precedence) or position
the value returned by a function is either the value specified in a call to
returnor the value of the last statement evaluated
Functions
Here is an example of a function that sums \(1,2,\dots,n\)
within the body of a function the arguments are referred to by their symbols and they take the value supplied at the time of invocation
any symbol found in the body of the function that does not match an argument has to be matched to a value by a process called scoping
Flow-control and operators
- R has all the standard flow control constructs that most computer langagues do
- if/else; while; repeat; for; break; next
- you can read the manual pages by calling
helpor using?(but for the latter you must quote the argument)
Logical Operators
- the short forms
&and|perform element-wise comparisons (vectorized) - the long forms
&&and||evaluate the first element only, move left to right and return when the result is determined - errors often occur when a programmer/analyst uses one form, when they want the other (R tries to warn you when it thinks there is a mistake)
Arithmetic Operators
- these are operators like
^or+ ?Syntaxwill get you the manual page for operator precedence- when in doubt always use parentheses - it is much clearer to the reader
Functions
in R functions are first class objects - this means they can be passed as arguments, assigned to symbols, stored in other data structures
in particular they can be passed as arguments to a function and returned as values
in some languages (e.g. C or Java) functions are not first class objects and they cannot be passed as arguments
Python uses a fairly similar strategy for functions to the one used in R (as do many other languages)
Scope
most of what computer languages do is map symbols (syntax) to values (semantics) and then create an executable program
when a computer comes upon an expression it parses it and that identifies the symbols that will need to be looked up
- the third part itself is a compound expression that can be decomposed into its parts,
*,bandc - in order to evaluate this the evaluator must find bindings for each of the symbols
- here we expect it wants numbers for
a,bandcand functions for+and*
##Scope
- all computer languages have a set of rules that are used to match these symbols to values
- one commonly used rule is to use lexical scope, but there are lots of different ways this is done
in
fun1(4)we probably all agree that the value that should be used forxis 4, we probably expect that+is a system functionbut what about
y- where should it’s value come from?
Scope
- Read the function below and try to understand what happens when it is evaluated:
Lexical Scope
lexical scope says that for any function with unbound values you should use the environment at the time the function was created to first look for bindings, in R (and Python) after that you look in the global environment (your workspace) and then in attached packages and system functions.
why is this useful?
Lexical Scope
we can write some interesting functions - like a function that evaluates the log likelihood for any given set of data
here we use
rexpto generate values from an Exponential distributionnotice that a function is returned and that R notes that it is a
closure
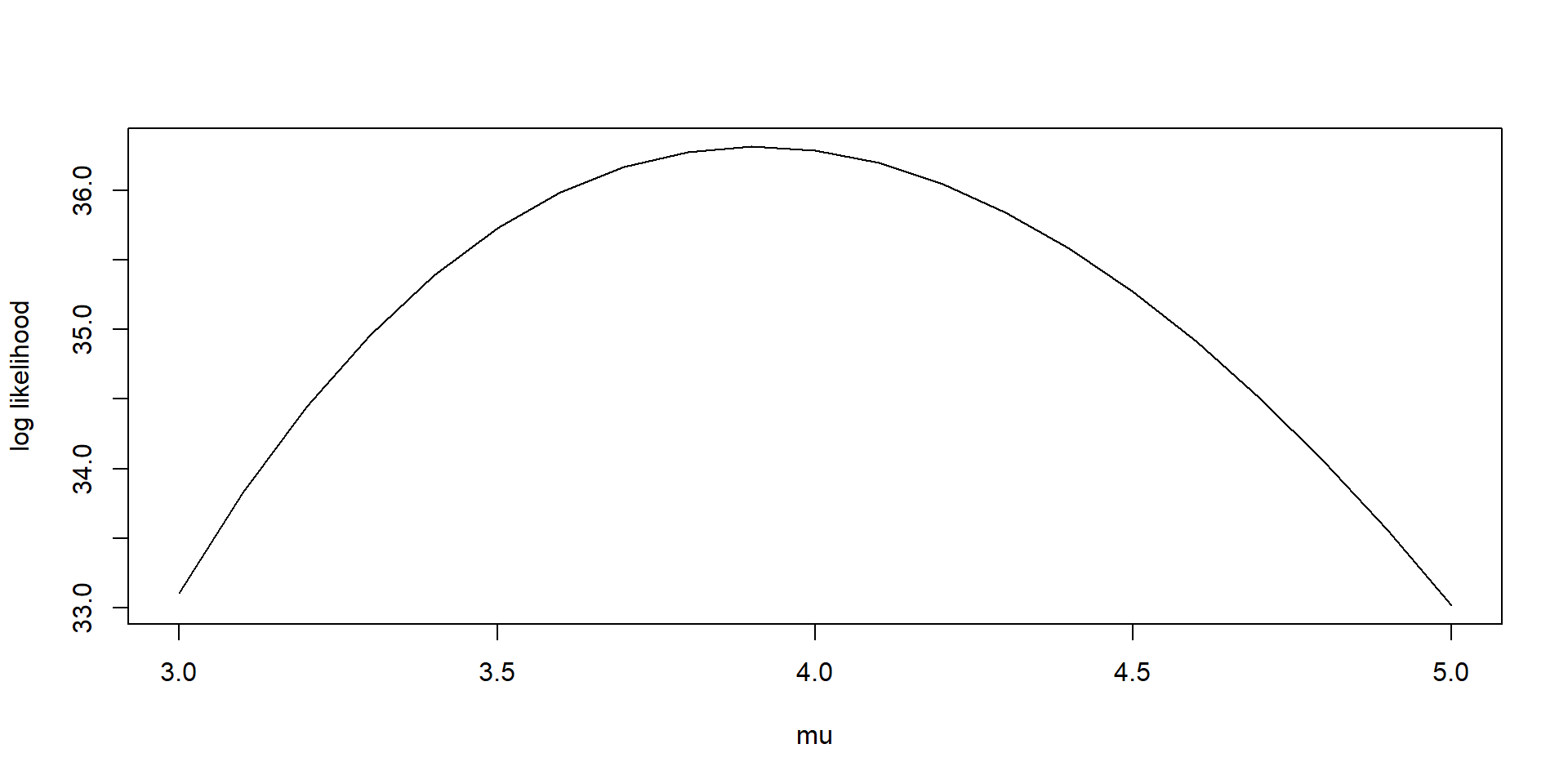
Lexical Scope Example
- here we generate different potential values for the rate parameter and plot the log likelihood - the MLE is the maximum of this
Name collisions
what happens when authors of two different packages choose the same name for their functions?
Look at how this function associated with the symbol
filterchanges in the following code segment:
by calling
library(dplyr)a new package has been put on the search list. You can callsearch()before and after the call tolibraryone implication of this observation is that users could inadvertently alter computations - and we will want to protect against that
Evaluation and the process used to find bindings
when evaluating a function R first establishes an evaluation environment that contains the formal arguments matched to the supplied arguments
if the function is in a package, then the Namespace directives of the package are used to augment this evaluation environment
any symbol not found in that evaluation environment will be searched for in the Global Environment (your workspace).
And after that the search path (
search()) in order.The evaluator will take the first match it finds and try to use that (sort of - it does know when it is looking for a function)
Namespaces
when authoring a package you will want to use Namespaces - the details will not be discussed here
If a package uses a Namespace then you can explicitly say which
filteryou want using:: :
Examples
- Restart your R Console and study this example:
Object Oriented Programming
R uses object oriented programming (OOP).
Base R uses two approaches referred to as S3 and S4, respectively.
S3, the original approach, is more common, but has some severe limitations
The S4 approach is more similar to the conventions used by the Lisp family of languages.
In S4 there are classes that are used to describe data structures and generic functions, that have methods associated with them
Object Oriented Programming
Object Oriented Programming
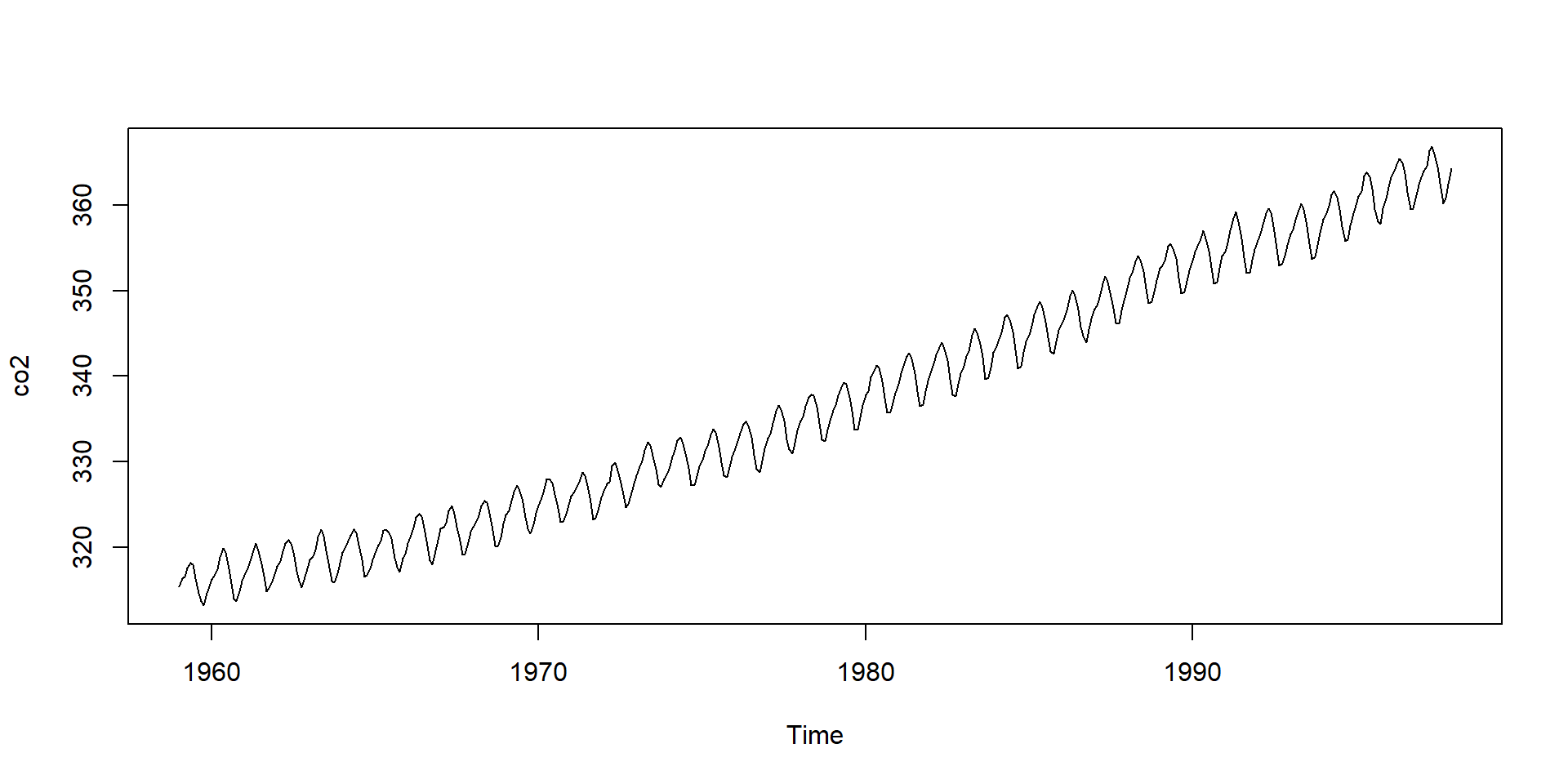
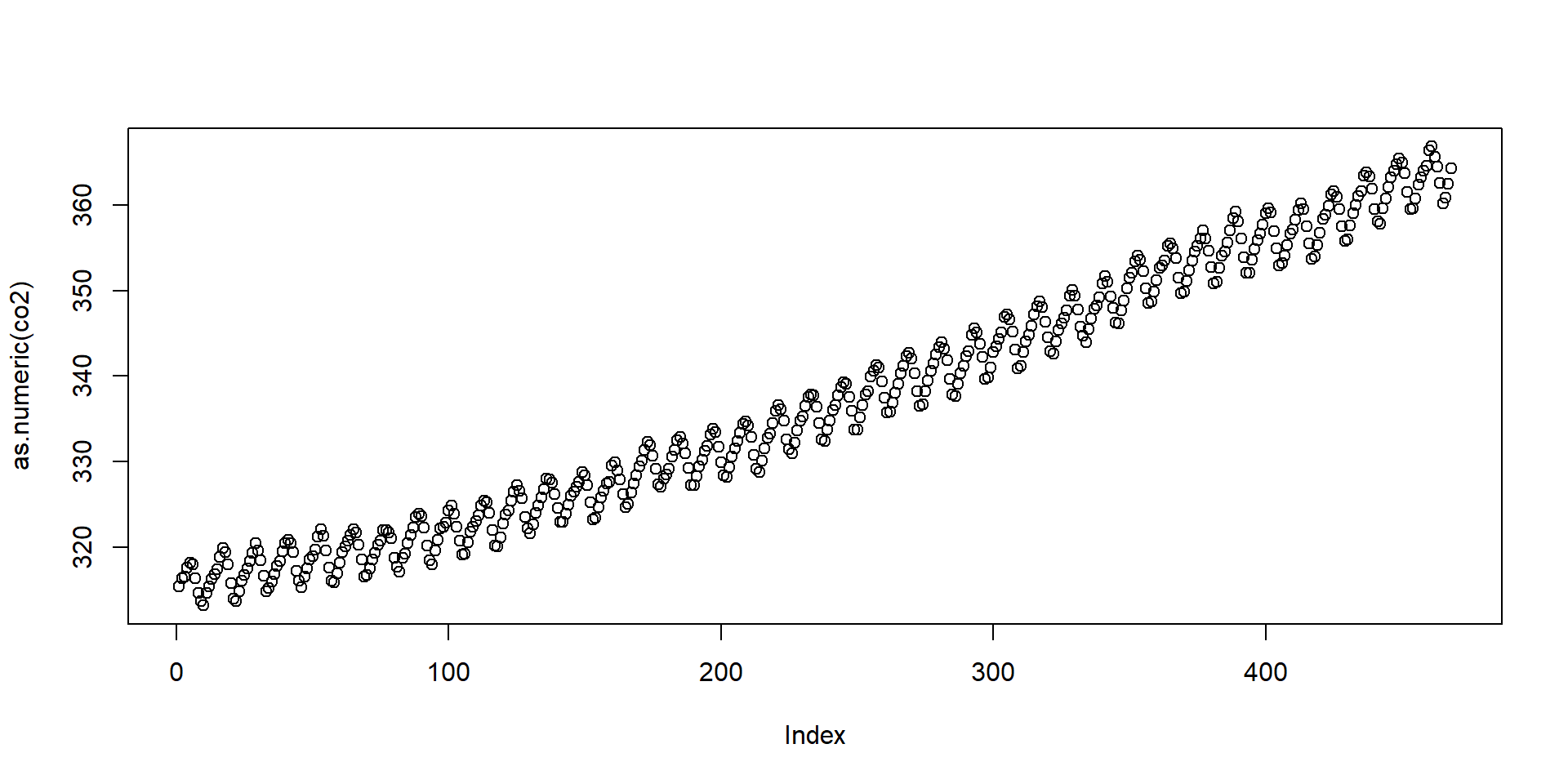
- Note
co2is not numeric:
- The plots are different because
plotbehaves differently with different classes.
Object Oriented Programming
- The first
plotactually calls the function
Notice all the plot functions that start with
plotby typingplot.and then tab.The function plot will call different functions depending on the class of the arguments.
Plots
Soon we will learn how to use the ggplot2 package to make plots.
R base does have functions for plotting though
Some you should know about are:
plot- mainly for making scatterplots.lines- add lines/curves to an existing plot.hist- to make a histogram.boxplot- makes boxplots.image- uses color to represent entries in a matrix.
Plots
Although, in general, we recommend using ggplot2, R base plots are often better for quick exploratory plots.
For example, to make a histogram of values in
xsimply type:
- To make a scatter plot of
yversusxand then interpolate we type: