library(tidyverse)
library(dslabs)
path <- system.file("extdata", package = "dslabs")
filename <- file.path(path, "fertility-two-countries-example.csv")
wide_data <- read_csv(filename)
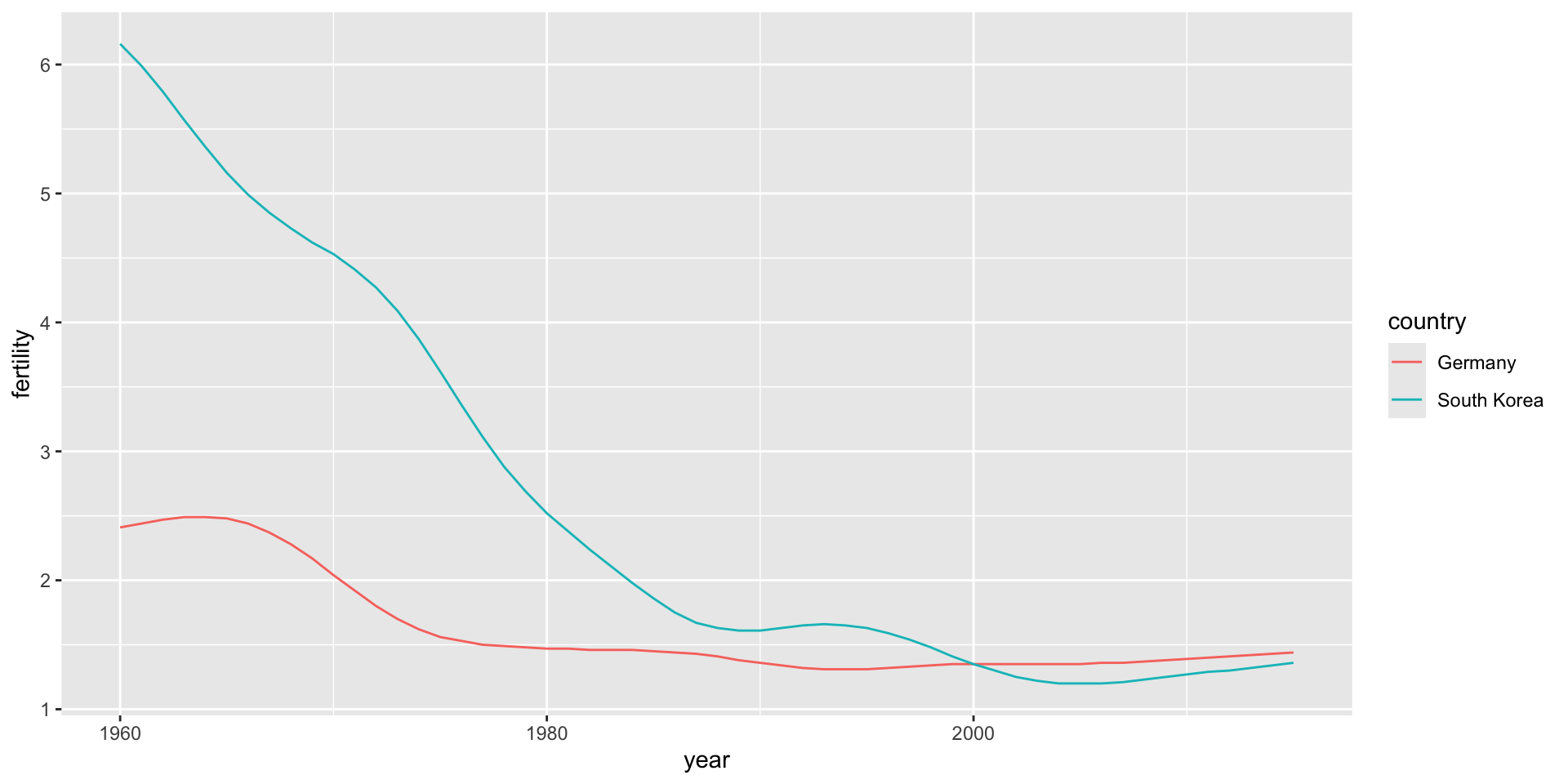
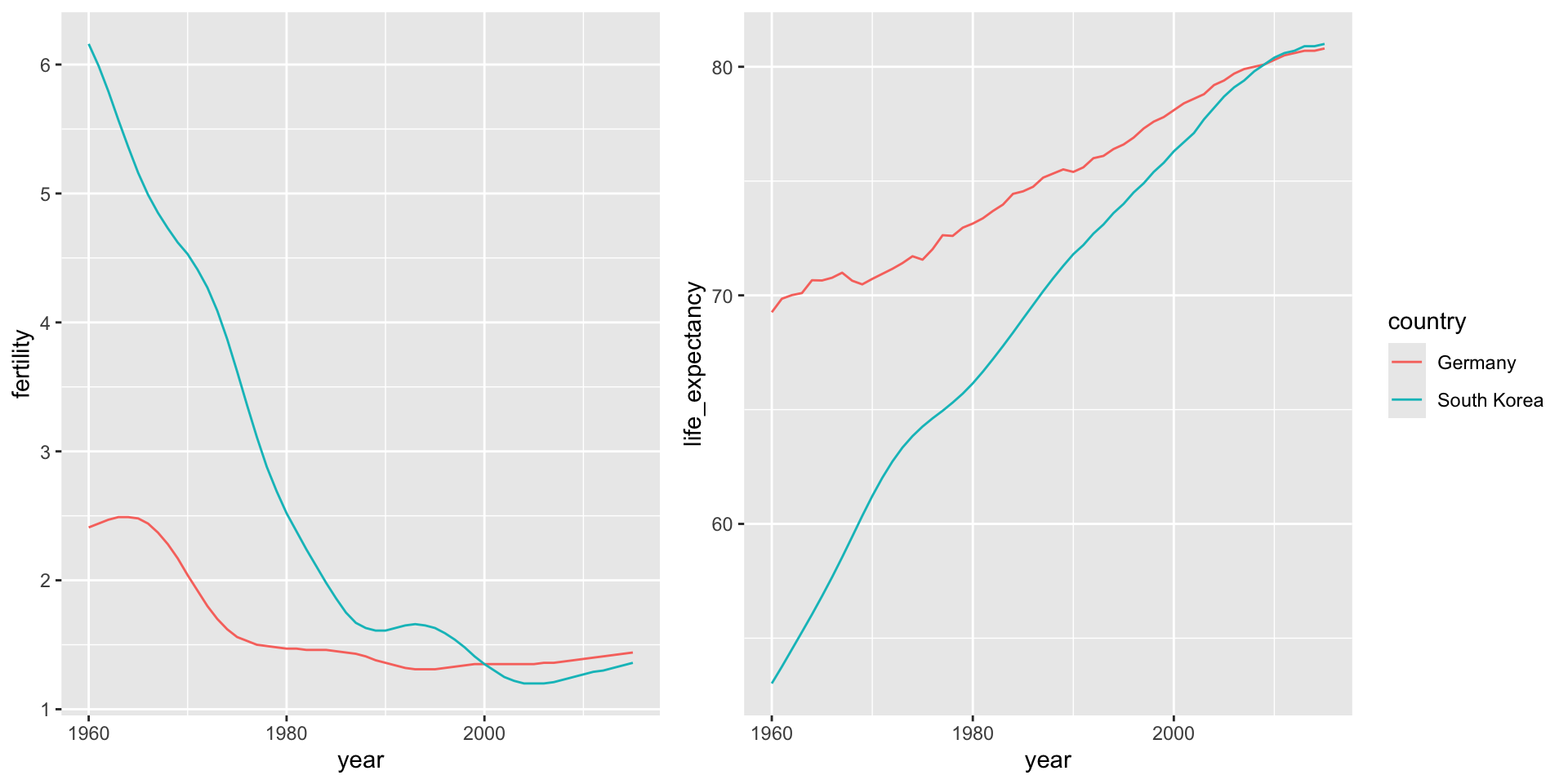
select(wide_data, 1:10)# A tibble: 2 × 10
country `1960` `1961` `1962` `1963` `1964` `1965` `1966` `1967` `1968`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Germany 2.41 2.44 2.47 2.49 2.49 2.48 2.44 2.37 2.28
2 South Korea 6.16 5.99 5.79 5.57 5.36 5.16 4.99 4.85 4.73