R Basics
2024-09-16
Packages
Use
install.packagesto install the dslabs package.Tryout the following functions:
sessionInfo,installed.packages
Prebuilt functions
Much of what we do in R is based on prebuilt functions.
Many are included in automatically loaded packages:
stats,graphics,grDevices,utils,datasets,methods.This subset of the R universe is refereed to as R base.
Very popular packages not included in R base:
ggplot2,dplyr,tidyr, anddata.table.
Important
For problem set 2 you can only use R base.
Prebuilt functions
Example of prebuilt functions that we will use today:
ls,rm,library,search,factor,list,exists,str,typeof, andclass.You can see the raw code for a function by typing it without the parenthesis: type
lson your console to see an example.
Help system
In R you can use
?orhelpto learn more about functions.You can learn about function using
or
The workspace
- Define a variable.
- Use
lsto see if it’s there. Also take a look at the Environment tab in RStudio.
- Use
rmto remove the variable you defined.
Variable name convention
A nice convention to follow is to use meaningful words that describe what is stored, use only lower case, and use underscores as a substitute for spaces.
For more we recommend this guide.
Data types
The main data types in R are:
One dimensional vectors: numeric, integer, logical, complex, characters.
Factors
Lists: this includes data frames.
Arrays: Matrices are the most widely used.
Date and time
tibble
S4 objects
Data types
Many errors in R come from confusing data types.
strstands for structure, gives us information about an object.typeofgives you the basic data type of the object. It reveals the lower-level, more fundamental type of an object in R’s memory.classThis function returns the class attribute of an object. The class of an object is essentiallytype_ofat a higher, often user-facing level.
Data types
Let’s see some example:
[1] "list"[1] "data.frame"'data.frame': 51 obs. of 5 variables:
$ state : chr "Alabama" "Alaska" "Arizona" "Arkansas" ...
$ abb : chr "AL" "AK" "AZ" "AR" ...
$ region : Factor w/ 4 levels "Northeast","South",..: 2 4 4 2 4 4 1 2 2 2 ...
$ population: num 4779736 710231 6392017 2915918 37253956 ...
$ total : num 135 19 232 93 1257 ...Data frames
Date frames are the most common class used in data analysis. It is like a spreadsheet.
Usually, rows represents observations and columns variables.
Each variable can be a different data type.
You can see part of the content like this
Data frames
- and all of the content like this:
- Type the above in RStudio.
Data frames
- A very common operation is adding columns like this:
state abb region population total pop_rank
1 Alabama AL South 4779736 135 29
2 Alaska AK West 710231 19 5
3 Arizona AZ West 6392017 232 36
4 Arkansas AR South 2915918 93 20
5 California CA West 37253956 1257 51
6 Colorado CO West 5029196 65 30Data frames
Note that we used
$.This is called the
accessorbecause it lets us access columns.
[1] 4779736 710231 6392017 2915918 37253956 5029196 3574097 897934
[9] 601723 19687653 9920000 1360301 1567582 12830632 6483802 3046355
[17] 2853118 4339367 4533372 1328361 5773552 6547629 9883640 5303925
[25] 2967297 5988927 989415 1826341 2700551 1316470 8791894 2059179
[33] 19378102 9535483 672591 11536504 3751351 3831074 12702379 1052567
[41] 4625364 814180 6346105 25145561 2763885 625741 8001024 6724540
[49] 1852994 5686986 563626- More generally: used to access components of a list.
Data frames
One way R confuses beginners is by having multiple ways of doing the same thing.
For example you can access the 4th column in the following five different ways:
- In general, we recommend using the name rather than the number as it is less likely to change.
with
withlet’s us use the column names as objects.This is convenient to avoid typing the data frame name over and over:
with
- Note you can write entire code chunks by enclosing it in curly brackets:
Vectors
- The columns of data frames are an example of one dimensional (atomic) vectors.
Vectors
Often we have to create vectors.
The concatenate function
cis the most basic way used to create vectors:
Sequences
- Sequences are a the common example of vectors we generate.
- When increasing by 1 you can use
:
Sequences
- A useful function to quickly generate the sequence
1:length(x)isseq_along:
- A reason to use this is to loop through entries:
Factors
One key distinction between data types you need to understad is the difference between factors and characters.
The
murderdataset has examples of both.
- Why do you think this is?
Factors
Factors store levels and the label of each level.
This is useful for categorical data.
Categories based on strata
In data analysis we often have to stratify continuous variables into categories.
The function
cuthelps us do this:
Categories based on strata
- We can assign it more meaningful level names:
age <- c(5, 93, 18, 102, 14, 22, 45, 65, 67, 25, 30, 16, 21)
cut(age, c(0, 11, 27, 43, 59, 78, 96, Inf),
labels = c("Alpha", "Zoomer", "Millennial", "X", "Boomer", "Silent", "Greatest")) [1] Alpha Silent Zoomer Greatest Zoomer Zoomer
[7] X Boomer Boomer Zoomer Millennial Zoomer
[13] Zoomer
Levels: Alpha Zoomer Millennial X Boomer Silent GreatestChanging levels
- This is often needed for ordinal data because R defaults to alphabetical order:
- You can change this with the
levelsargument:
Changing levels
A common reason we need to change levels is to assure R is aware which is the reference strata.
This is important for linear models because the first level is assumed to be the reference.
Changing levels
We often want to order strata based on a summary statistic.
This is common in data visualization.
We can use
reorderfor this:
Factors
- Another reason we used factors is because they more efficient:
80000232 bytes40000648 bytes- An integer is easier to store than a character string.
Factors
Exercise: How can we make this go much faster?
Factors can be confusing
- Try to make sense of this:
Factors can be confusing
- Avoid keeping extra levels with
droplevels:
- But note what happens if we change to another level:
NAs
NA stands for not available.
Data analysts have to deal with NAs often.
NAs
- dslabs includes an example dataset with NAs
- The
is.nafunction is key for dealing with NAs
NAs
- Technically NA is a logical
- When used with ands and ors, NAs behaves like FALSE
- But NA is not FALSE. Try this:
NaNs
A related constant is
NaN.Unlike
NA, which is a logical,NaNis a number.It is a
numericthat is Not a Number.Here are some examples:
Coercing
When you do something inconsistent with data types, R tries to figure out what you mean and change it accordingly.
We call this coercing.
R does not return an error and in some cases does not return a warning either.
This can cause confusion and unnoticed errors.
So it’s important to understand how and when it happens.
Coercing
- Here are some examples:
Coercing
- When R can’t figure out how to coerce, rather an error it returns an NA:
- Note that including
NAs in arithmetical operations usually returns anNA.
Coercing
You want to avoid automatic coercion and instead explicitly do it.
Most coercion functions start with
as.Here is an example.
Coercing
- More examples:
Lists
Data frames are a type of list.
Lists permit components of different types and, unlike data frames, different lengths:
- The JSON format is best represented as list in R.
Lists
- You can access components in different ways:
Matrics
Matrices are another widely used data type.
They are similar to data frames except all entries need to be of the same type.
We will learn more about matrices in the High Dimensional data Analysis part of the class.
Functions
- You can define your own function. The form is like this:
Functions
- Here is an example of a function that sums \(1,2,\dots,n\)
Lexical scope
- Study what happens here:
Namespaces
- Look at how this function changes by typing the following:
Namespaces
Note what R searches the Global Environment first.
Use
searchto see other environments R searches.Note many prebuilt functions are in
stats.
Namespaces
- You can explicitly say which
filteryou want using namespaces:
Namespaces
- Restart yoru R Consuole and study this example:
Object Oriented Programming
R uses object oriented programming (OOP).
It uses two approaches referred to as S3 and S4, respectively.
S3, the original approach, is more common.
The S4 approach is more similar to the conventions used by modern OOP languages.
Object Oriented Programming
Object Oriented Programming
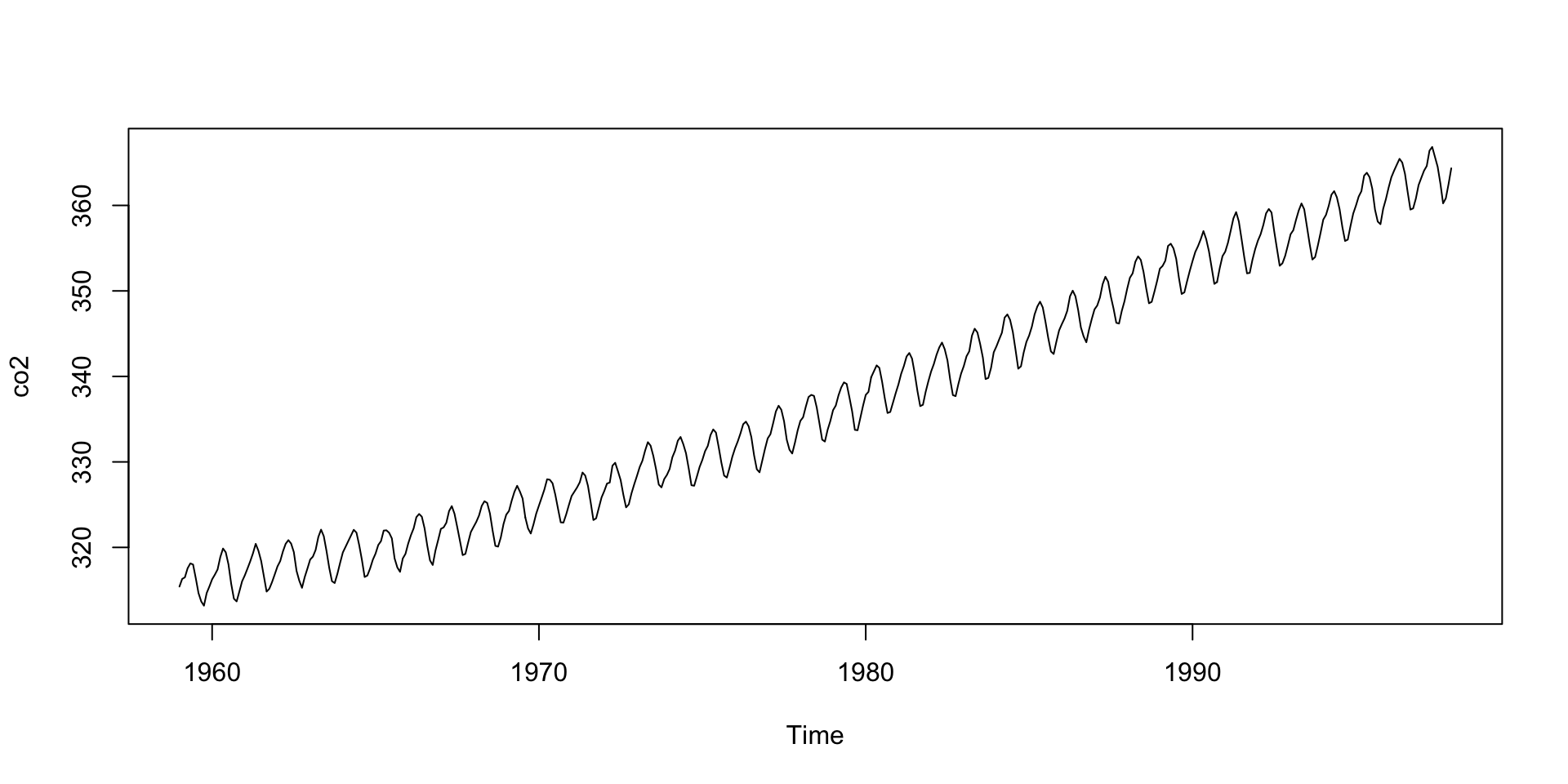
- Note
co2is not numeric:
- The plots are different because
plotbehaves different with different classes.
Object Oriented Programming
- The first
plotactually calls the function
Notice all the plot functions that start with
plotby typingplot.and then tab.The function plot will call different functions depending on the class of the arguments.
Plots
Soon we will learn how to use the ggplot2 package to make plots.
R base does have functions for plotting though
Some you should know about are:
plot- mainly for making scatterplots.lines- add lines/curves to an existing plot.hist- to make a histogram.boxplot- makes boxplots.image- uses color to represent entries in a matrix.
Plots
Although, in general, we recommend using ggplot2, R base plots are often better for quick exploratory plots.
For example, to make a histogram of values in
xsimply type:
- To make a scatter plot of
yversusxand then interpolate we type: